Information accessibility snRNA-seq information are offered in Gene Expression Omnibus (GEO) under the accession 19459007 GSE185472 19459008 The following public datasets were utilized for snRNA-seq analysis: Allen Brain Institute human adult snRNA-seq information from numerous cortical locations ( https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq 19459008; accessed October 2022), snRNA-seq information from broad temporal protection from fetal to the adult years phases of the Brodmann location 8, 9, 10 and 46 prefrontal cortex areas (GEO accession GSE168408 19459008 and snRNA-seq from 8-month-old cortical organoid transplants (GEO accession GSE190815. For single-nucleus analysis, we utilized hg19 human referral genome v1.2.0 and mm10 mouse recommendation genome v1.2.0 offered by 10x Genomics. The series and gene files utilized to construct the referrals can be accomplished at 19459012 ftp:// ftp.ensembl.org/pub/grch37/release-84/fasta/homo_sapiens/dna/ 19459008 and ftp:// ftp.ensembl.org/pub/grch37/release-84/gtf/homo_sapiens/ 19459008 (for human hg19 genome); 19459014 ftp:// ftp.ensembl.org/pub/release-84/fasta/mus_musculus/dna/ 19459008 and 19459015 ftp:// ftp.ensembl.org/pub/release-84/gtf/mus_musculus/ (for mouse mm10 genome). All other raw information utilized for outlining in the figures are offered as source information. 19459016 Source information are offered with this paper. Referrals Liddelow, S. A. et al. Neurotoxic reactive astrocytes are caused by triggered microglia. Nature 541 19459028, 481– 487 (2017 ). Post 19459030 ADS 19459008 19459031 CAS 19459032 PubMed 19459033 PubMed Central Google Scholar 19459008 19659007 Qian, X., Song, H. & & Ming, G. L. Brain organoids: advances, applications and difficulties. Advancement 146 , dev166074 (2019 ). Post 19459008 19459039 CAS 19459040 PubMed Central 19459008 Google Scholar Wang, M., Zhang, L. & & Gage, F. H. Modeling neuropsychiatric conditions utilizing human caused pluripotent stem cells. Protein Cell 19659011 11 19459028, 45– 59 (2020 ). Post 19459046 CAS 19459008 PubMed 19459048 Google Scholar Lancaster, M. A. et al. Cerebral organoids design human brain advancement and microcephaly. 19459025 Nature 501 19459028, 373– 379 (2013 ). Short article 19459052 ADS 19459053 CAS PubMed 19459055 Google Scholar 19659016 Kim, J., Koo, B. K. & & Knoblich, J. A. Human organoids: design systems for human biology and medication. Nat. Rev. Mol. Cell Biol. 21 , 571– 584 (2020 ). 19659018 Post 19459059 CAS 19459008 19459060 PubMed 19459061 PubMed Central 19459008 19459062 Google Scholar 19659019 Velasco, S. et al. Private brain organoids reproducibly form cell variety of the human cortex. 19459025 Nature 19659020 570 , 523– 527 (2019 ). Short article 19459066 ADS 19459008 CAS 19459008 PubMed 19459008 19459069 PubMed Central 19459008 19459070 Google Scholar 19459008 19659022 Sloan, S. A. et al. Human astrocyte maturation recorded in 3D cerebral cortical spheroids originated from pluripotent stem cells. 19459025 Nerve cell 95 19459028, 779– 790 (2017 ). Post CAS 19459075 PubMed 19459008 19459076 PubMed Central 19459077 Google Scholar 19659025 Qian, X. et al. Sliced up human cortical organoids for modeling unique cortical layer development. 19459025 Cell Stem Cell 19659026 26 , 766– 781 (2020 ). Short article CAS PubMed Central 19459084 Google Scholar 19459008 19659028 Molofsky, A. V. et al. Astrocytes and illness: a neurodevelopmental point of view. 19459025 Genes Dev. 26 , 891– 907 (2012 ). Post 19459008 19459088 CAS 19459008 19459089 PubMed Central 19459091 Google Scholar Tchieu, J. et al. NFIA is a gliogenic switch making it possible for fast derivation of practical human astrocytes from pluripotent stem cells. 19459025 Nat. Biotechnol. 19659032 37 , 267– 275 (2019 ). Short article 19459095 CAS 19459096 PubMed 19459008 PubMed Central 19459098 Google Scholar Canals, I. et al. Quick and effective induction of practical astrocytes from human pluripotent stem cells. 19459025 Nat. Approaches 19659035 15 , 693– 696 (2018 ). 19659036 Post 19459008 19459102 CAS 19459008 PubMed 19459008 Google Scholar 19659037 Santos, R. et al. Distinction of inflammation-responsive astrocytes from glial progenitors created from human induced pluripotent stem cells. Stem Cell Reports 8 19459028, 1757– 1769 (2017 ). Short article 19459108 CAS 19459109 PubMed 19459008 PubMed Central Google Scholar 19459008 19659040 Barbar, L. et al. CD49f is an unique marker of practical and reactive human iPSC-derived astrocytes. 19459025 Nerve cell 19659041 107 , 436– 453 (2020 ). Short article 19459008 CAS PubMed 19459008 PubMed Central Google Scholar 19459008 Zhang, J. & & Liu, Q. Cholesterol metabolic process and homeostasis in the brain. 19459025 Protein Cell 19659044 6 , 254– 264 (2015 ). 19659045 Short article CAS PubMed 19459124 PubMed Central 19459125 Google Scholar 19459008 19659046 Daneman, R. & & Prat, A. The blood-brain barrier. 19459025 Cold Spring Harb. Perspect. Biol. 7 19459028, a020412 (2015 ). Short article PubMed 19459130 PubMed Central Google Scholar 19459008 19659049 Mansour, A. A. et al. An in vivo design of practical and vascularized human brain organoids. 19459025 Nat. Biotechnol. 36 19459028, 432– 441 (2018 ). 19659051 Post 19459135 CAS PubMed 19459008 19459137 PubMed Central 19459138 Google Scholar 19459008 19659052 Bao, Z. et al. Human cerebral organoid implantation eased the neurological deficits of distressing brain injury in mice. Oxid. Medication. Cell. Longev. 2021 19459028, 6338722 (2021 ). 19659054 Short article 19459008 PubMed 19459008 19459143 PubMed Central 19459144 Google Scholar 19659055 Daviaud, N., Friedel, R. H. & & Zou, H. Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. 19459025 eNeuro 5 19459028, ENEURO.0219-18.2018 (2018 ). Short article 19459148 PubMed 19459008 PubMed Central 19459008 Google Scholar 19659058 Kitahara, T. et al. Axonal extensions along corticospinal systems from transplanted human cerebral organoids. Stem Cell Reports 19659059 15 19459028, 467– 481 (2020 ). Short article 19459154 CAS PubMed 19459008 19459156 PubMed Central 19459157 Google Scholar Shi, Y. et al. Vascularized human cortical organoids (vOrganoids) design cortical advancement in vivo. PLoS Biol. 18 19459028, e3000705 (2020 ). 19659063 Post 19459161 CAS 19459008 19459162 PubMed 19459008 PubMed Central 19459164 Google Scholar 19459008 19659064 Revah, O. et al. Maturation and circuit combination of transplanted human cortical organoids. Nature 19659065 610 , 319– 326 (2022 ). 19659066 Short article 19459008 ADS 19459008 CAS 19459008 19459170 PubMed 19459171 PubMed Central 19459008 Google Scholar 19459008 19659067 Jgamadze, D. et al. Structural and practical combination of human forebrain organoids with the hurt adult rat visual system. 19459025 Cell Stem Cell 19659068 30 , 137– 152 (2023 ). Post 19459176 CAS 19459008 PubMed 19459178 PubMed Central 19459008 Google Scholar Qian, X. et al. Generation of human brain region-specific organoids utilizing a miniaturized spinning bioreactor. Nat. Protoc. 19659071 13 19459028, 565– 580 (2018 ). Post 19459008 19459183 CAS PubMed 19459008 PubMed Central Google Scholar 19459008 Qian, X. et al. Brain-region-specific organoids utilizing mini-bioreactors for modeling ZIKV direct exposure. 19459025 Cell 165 19459028, 1238– 1254 (2016 ). 19659075 Short article 19459190 CAS 19459008 19459191 PubMed 19459192 PubMed Central Google Scholar 19459008 19659076 Langfelder, P. & & Horvath, S. WGCNA: an R plan for weighted connection network analysis. BMC Bioinformatics 19659077 9 19459028, 559 (2008 ). Short article PubMed 19459008 PubMed Central Google Scholar Morabito, S. et al. Single-nucleus chromatin ease of access and transcriptomic characterization of Alzheimer’s illness. 19459025 Nat. Genet. 19659080 53 19459028, 1143– 1155 (2021 ). Post CAS PubMed 19459205 PubMed Central 19459008 Google Scholar 19659082 Allen, N. J. & & Eroglu, C. Cell biology of astrocyte-synapse interactions. Nerve cell 96 19459028, 697– 708 (2017 ). 19659084 Post CAS 19459008 PubMed 19459008 PubMed Central 19459008 19459213 Google Scholar Stogsdill, J. A. et al. Astrocytic neuroligins manage astrocyte morphogenesis and synaptogenesis. 19459025 Nature 551 19459028, 192– 197 (2017 ). 19659087 Post 19459217 ADS 19459218 CAS 19459219 PubMed 19459008 PubMed Central Google Scholar Oberheim, N. A. et al. Distinctively hominid functions of adult human astrocytes. J. Neurosci. 19659089 29 19459028, 3276– 3287 (2009 ). 19659090 Short article CAS 19459226 PubMed 19459008 PubMed Central 19459228 Google Scholar 19459008 Zhang, Y. et al. Filtration and characterization of progenitor and fully grown human astrocytes exposes transcriptional and practical distinctions with mouse. Nerve cell 19659092 89 , 37– 53 (2016 ). Post 19459008 19459232 CAS 19459233 PubMed Google Scholar 19459008 19659094 Oberheim, N. A., Wang, X., Goldman, S. & & Nedergaard, M. Astrocytic intricacy differentiates the human brain. 19459025 Patterns Neurosci. 19659095 29 , 547– 553 (2006 ). 19659096 Short article CAS 19459008 PubMed 19459240 Google Scholar Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in repaired tissue. Nat. Biotechnol. 38 , 586– 599 (2020 ). 19659099 Post 19459008 19459244 CAS 19459008 19459245 PubMed Google Scholar 19659100 Li, Y. et al. Direct labeling and visualization of capillary with lipophilic carbocyanine color DiI. 19459025 Nat. Protoc. 19659101 3 19459028, 1703– 1708 (2008 ). Post 19459008 CAS 19459008 PubMed 19459008 19459252 PubMed Central Google Scholar Herring, C. A. et al. Human prefrontal cortex gene regulative characteristics from pregnancy to the adult years at single-cell resolution. 19459025 Cell 185 19459028, 4428– 4447 (2022 ). 19659105 Short article 19459008 19459257 CAS PubMed 19459259 Google Scholar Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566 19459028, 496– 502 (2019 ). Short article 19459008 ADS 19459264 CAS 19459265 PubMed 19459266 PubMed Central 19459267 Google Scholar 19659109 Sofroniew, M. V. & & Vinters, H. V. Astrocytes: biology and pathology. 19459025 Acta Neuropathol. 119 , 7– 35 (2010 ). Post 19459271 PubMed Google Scholar Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 19659113 32 19459028, 6391– 6410 (2012 ). Post CAS PubMed 19459008 PubMed Central 19459008 Google Scholar Anderson, M. A. et al. Astrocyte scar development help main nerve system axon regrowth. Nature 19659116 532 19459028, 195– 200 (2016 ). Short article ADS 19459008 CAS 19459285 PubMed 19459008 19459286 PubMed Central 19459008 19459287 Google Scholar 19459008 19659118 Tarrago, M. G. et al. A powerful and particular CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD 19459290 + decrease. Cell Metab. 27 19459028, 1081– 1095 e1010 (2018 ). Short article CAS 19459008 PubMed 19459295 PubMed Central Google Scholar 19659121 Sprenger, H. G. & & Langer, T. The excellent and the bad of mitochondrial separations. Patterns Cell Biol. 29 , 888– 900 (2019 ). Short article CAS 19459301 PubMed 19459008 19459302 Google Scholar Krencik, R. & & Zhang, S. C. Directed distinction of practical astroglial subtypes from human pluripotent stem cells. 19459025 Nat. Protoc. 6 19459028, 1710– 1717 (2011 ). Post 19459306 CAS PubMed 19459008 19459308 PubMed Central 19459008 19459309 Google Scholar 19459008 19659127 Palm, T. et al. Fast and robust generation of long-lasting self-renewing human neural stem cells with the capability to create fully grown astroglia. 19459025 Sci. Rep. 5 19459028, 16321 (2015 ). Short article ADS 19459008 19459314 CAS 19459008 19459315 PubMed 19459316 PubMed Central 19459008 19459317 Google Scholar Han, X. et al. Forebrain engraftment by human glial progenitor cells boosts synaptic plasticity and knowing in adult mice. 19459025 Cell Stem Cell 19659131 12 19459028, 342– 353 (2013 ). 19659132 Short article CAS PubMed 19459008 19459323 PubMed Central 19459008 Google Scholar 19659133 Windrem, M. S. et al. A competitive benefit by neonatally engrafted human glial progenitors yields mice whose brains are chimeric for human glia. 19459025 J. Neurosci. 34 , 16153– 16161 (2014 ). 19659135 Post 19459008 19459328 PubMed 19459008 19459329 PubMed Central 19459008 Google Scholar 19459008 19659136 Mariani, J. N., Zou, L. & & Goldman, S. A. Human glial chimeric mice to specify the function of glial pathology in human illness. 19459025 Techniques Mol. Biol. 19659137 1936 , 311– 331 (2019 ). 19659138 Post 19459008 19459334 CAS PubMed Central 19459008 19459337 Google Scholar 19659139 Zeisel, A. et al. Brain structure. Cell enters the mouse cortex and hippocampus exposed by single-cell RNA-seq. Science 347 19459028, 1138– 1142 (2015 ). Post 19459008 ADS 19459008 CAS 19459008 PubMed 19459008 19459344 Google Scholar 19459008 Bayraktar, O. A. et al. Astrocyte layers in the mammalian cortex exposed by a single-cell in situ transcriptomic map. Nat. Neurosci. 19659143 23 , 500– 509 (2020 ). 19659144 Post CAS 19459008 PubMed 19459008 PubMed Central 19459008 Google Scholar 19459008 Hodge, R. D. et al. Saved cell types with divergent functions in human versus mouse cortex. 19459025 Nature 573 , 61– 68 (2019 ). 19659147 Short article 19459008 19459355 ADS 19459008 CAS 19459357 PubMed Central 19459359 Google Scholar 19459008 Jorstad, N. L. et al. Transcriptomic cytoarchitecture exposes concepts of human neocortex company. 19459025 Science 382 , eadf6812 (2023 ). Post 19459008 CAS PubMed 19459008 19459365 Google Scholar 19459008 19659151 Burda, J. E. et al. Divergent transcriptional policy of astrocyte reactivity throughout conditions. Nature 606 19459028, 557– 564 (2022 ). Post 19459008 19459369 ADS 19459008 19459370 CAS 19459008 19459371 PubMed 19459008 19459372 PubMed Central Google Scholar 19659154 Cowan, C. A. et al. Derivation of embryonic stem-cell lines from human blastocysts. New Engl. J. Med. 19659155 350 , 1353– 1356 (2004 ). 19659156 Post 19459377 CAS 19459378 PubMed 19459008 Google Scholar 19459008 19659157 Thomson, J. A. et al. Embryonic stem cell lines stemmed from human blastocysts. 19459025 Science 282 19459028, 1145– 1147 (1998 ). Short article 19459383 ADS 19459008 CAS 19459008 PubMed Google Scholar Mertens, J. et al. Differential reactions to lithium in hyperexcitable nerve cells from clients with bipolar illness. 19459025 Nature 527 19459028, 95– 99 (2015 ). Post CAS PubMed Central 19459008 19459393 Google Scholar Goncalves, J. T. et al. In vivo imaging of dendritic pruning in dentate granule cells. Nat. Neurosci. 19 , 788– 791 (2016 ). Short article 19459397 MathSciNet 19459008 CAS 19459399 PubMed 19459400 PubMed Central 19459401 Google Scholar 19659166 Longair, M. H., Baker, D. A. & & Armstrong, J. D. Simple Neurite Tracer: open source software application for restoration, visualization and analysis of neuronal procedures. Bioinformatics 19659167 27 , 2453– 2454 (2011 ). Short article 19459008 CAS PubMed Google Scholar 19659169 Kuwajima, M., Mendenhall, J. M. & & Harris, K. M. Large-volume restoration of brain tissue from high-resolution serial area images obtained by SEM-based scanning transmission electron microscopy. 19459025 Approaches Mol. Biol. 950 , 253– 273 (2013 ). Post CAS 19459412 PubMed 19459413 PubMed Central 19459414 Google Scholar 19459008 19659172 Deerinck, T. J., Bushong, E. A., Thor, A. & & Ellisman, M. H. NCMIR techniques for 3D EM: a brand-new procedure for preparation of biological specimens for serial block face scanning electron microscopy. 19459025 Microscopy 1 , 6– 8 (2010 ). 19659174 Google Scholar Horstmann, H., Korber, C., Satzler, K., Aydin, D. & & Kuner, T. Serial area scanning electron microscopy (S3EM) on silicon wafers for ultra-structural volume imaging of cells and tissues. PLoS ONE 7 , e35172 (2012 ). 19659177 Post 19459008 19459421 ADS CAS 19459008 PubMed 19459424 PubMed Central 19459425 Google Scholar Hao, Y. et al. Integrated analysis of multimodal single-cell information. 19459025 Cell 184 , 3573– 3587 e3529 (2021 ). 19659180 Post 19459008 19459429 CAS 19459008 PubMed 19459008 19459431 PubMed Central 19459008 19459432 Google Scholar Conway, J. R., Lex, A. & & Gehlenborg, N. UpSetR: an R bundle for the visualization of converging sets and their residential or commercial properties. 19459025 Bioinformatics 33 , 2938– 2940 (2017 ). 19659183 Post 19459008 19459436 CAS PubMed 19459008 19459438 PubMed Central 19459008 19459439 Google Scholar 19459008 Kuleshov, M. V. et al. Enrichr: an extensive gene set enrichment analysis web server 2016 upgrade. 19459025 Nucleic Acids Res. 19659185 44 , W90– W97 (2016 ). Short article 19459008 19459443 CAS 19459008 19459444 PubMed Central Google Scholar Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z. & & Zhang, B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. 19459025 Nucleic Acids Res. 19659188 47 19459028, W199– W205 (2019 ). Post CAS 19459008 PubMed Central 19459008 Google Scholar 19459008 Download referrals 19659191 Recognitions We thank A. Mansour (Salk Institute) for offering the pCSC-CAG-GFP lentiviral identified H9 ESC line, N. Hah for technical help with snRNA-seq and R. Garg for support with EM division. We likewise thank M.L. Gage for editorial remarks and A. Cao and T. Bartol for their assist with 3DEM visualization. This work was supported by the American Heart Association and the Paul G. Allen Frontiers Group Grant (19PABHI34610000/TEAM LEADER: Fred H. Gage/2019), JPB Foundation, Annette C. Merle-Smith, Lynn and Edward Streim, the Milky Way Foundation, the Ray and Dagmar Dolby Family Fund and NIH (R37 AG072502-03, P30 AG062429-05, P30 AG068635-04, R01 AG070154-04, AG056306-07 and P01 AG051449-08). This work was likewise supported by the NGS Core Facility, the GT3 Core Facility, the Razavi Newman Integrative Genomics and Bioinformatics Core Facility and the Waitt Advanced Biophotonics Core Facility of the Salk Institute with financing from NIH– NCI (CCSG, P30 014195), the Chapman Foundation, NINDS R24 Core Grant, NEI and the Waitt Foundation. M.W. is supported by a Young Investigator Grant from the Brain & & Behavior Research Foundation (BBRFNARSAD) and a Pioneer Fund Postdoctoral Scholar Award from the Salk Institute. Figures 19459458 2a 19459008, 2h , 19459459 4a 19459008, 5a , 6a and 19459461 6d 19459008 were developed with 19459462 BioRender.com 19459008 19659193 Author info 19659194 Author notes Uri Manor 19659196 Present address: Division of Biological Sciences, University of California, San Diego, La Jolla, CA, USA 19659197 These authors contributed similarly: Meiyan Wang, Lei Zhang. Authors and Affiliations Lab of Genetics, The Salk Institute for Biological Studies, La Jolla, CA, USA Meiyan Wang, Lei Zhang, Iryna S. Gallina, Lynne L. Xu, Christina K. Lim, Sarah Fernandes, Monisha D. Saxena, Shashank Coorapati, Sarah L. Parylak & & Fred H. Gage Waitt Advanced Biophotonics Core, The Salk Institute for Biological Studies, La Jolla, CA, USA Sammy Weiser Novak, Leonardo R. Andrade & & Uri Manor 19659203 Integrative Genomics and Bioinformatics Core, The Salk Institute for Biological Studies, La Jolla, CA, USA Jingting Yu, Maxim N. Shokhirev & & April E. Williams Department of Biological Sciences, University of California, San Diego, La Jolla, CA, USA 19659206 Christina K. Lim & & Sarah Fernandes 19659207 Next Generation Sequencing Core, The Salk Institute for Biological Studies, La Jolla, CA, USA 19659208 Cristian Quintero & & Elsa Molina 19659209 Contributions M.W., L.Z. and F.H.G. developed the research study and composed the paper. M.W., L.Z., C.K.L. and S.F. carried out cell culture and organoid distinction. M.W., L.Z., C.K.L. and I.S.G. carried out surgical treatments. S.L.P. helped in surgical treatments. M.W. and I.S.G. carried out snRNA-seq and scRNA-seq. M.W., C.Q. and E.M. carried out NanoString GeoMx DSP. M.W., M.N.S., J.Y. and A.E.W. carried out bioinformatics analyses. M.W. and L.Z. carried out imaging analysis with support from L.L.X., C.K.L., S.C. and M.D.S. L.Z., S.W.N. and L.R.A. carried out sample processing for SEM. S.W.N. and L.R.A. carried out electron tiny image analysis under the guidance of M.W., L.Z. and U.M. F.H.G. offered financing. 19659211 Corresponding author 19659212 Correspondence to Fred H. Gage Principles statements 19659214 Completing interests 19659215 The authors state no contending interests. Peer evaluation 19659217 Peer evaluation details 19659218 Nature Biotechnology 19459026 thanks the confidential customers for their contribution to the peer evaluation of this work. 19659219 Extra info 19659220 Publisher’s note 19459028 Springer Nature stays neutral with regard to jurisdictional claims in released maps and institutional associations. 19659221 Prolonged information 19659222 Extended Data Fig. 1 Rapid astrogliogenesis in glia-enriched cortical organoids. a Test bright-field pictures of hiPSCs/hESCs nests and glia-enriched cortical organoids at days 0, 14, 30 and 60. Day 0 describes embryoid bodies (EB). Scale bars, 890 μm. 19459027 b Quantitative PCR analysis of the expression levels of NFIA and SOX2 in three-week-old organoids cultured under differing conditions (Hues6: 1, n= 5; 2, n= 5; 3, n= 5; 4, n= 5; 5, n= 4. iPSC822: 1, n= 5; 2, n= 5; 3, n= 5; 4, n= 5; 5, n= 4). Each dot represents a swimming pool of 3 organoids. Bars, suggest ± s.d. Two-sided t-test, ns, not considerable, * p < 0.05, *** p < 0.001, **** p < 0.0001. 19459027 c Immunostaining of two-month-old glia-enriched cortical organoids: stem cells (SOX2, magenta), astrocytes (GFAP, green). Scale bar, 100 μm. d Immunostaining of two-month-old glia-enriched cortical organoids: intermediate progenitor cells (EOMES, green), cortical excitatory nerve cells (CTIP2, magenta; SATB2, gray), stem cells (SOX2, green), nerve cells (NeuN, magenta), astrocytes (GFAP, gray), glia (HOPX, green; S100B, magenta; GFAP, gray). Scale bars, 20 μm. 19459027 e 19459028 Immunostaining of three-month-old glia-enriched cortical organoids distinguished in 2% FBS (leading left panel), 2% SATO (leading best panel; serum-free condition, SATO part reported in ref. 30 , 10% FBS (bottom left panel) and 10% SATO (bottom ideal panel). Astrocytes (GFAP, green). Scale bars, 100 μm. 19459027 f Metrology of the GFAP fluorescence strength in glia-enriched cortical organoids distinguished in 2% FBS, 2% SATO, 10% FBS and 10% SATO conditions (Hues6: 10% FBS, n= 3; 10% SATO, n= 6; 2% FBS, n= 4; 2% SATO, n= 6. iPSC822: 10% FBS, n= 6; 10% SATO, n= 8; 2% FBS, n= 5; 2% SATO, n= 6). Each dot represents one organoid. Bars, imply ± s.e.m. Two-sided t-test, *** p < 0.001, **** p < 0.0001. 19659226 Source information Extended Data Fig. 2 snRNA-seq of 10-week-old glia-enriched cortical organoids. a UMAP plot of snRNA-seq information from 10-week-old glia-enriched cortical organoids. The portion of each significant cell types is revealed. APC, astrocyte progenitor cell; Ast, astrocyte; IPC, intermediate progenitor cell; In, repressive nerve cell; Cortical Ex, cortical excitatory nerve cell; Ex1 and Ex2, excitatory nerve cell. 19459027 b 19459028 Expression of picked marker genes utilized in cell type recognition. The violin plot reveals the circulation of stabilized expression in nuclei in each cluster. Scale: stabilized checked out counts. 19459027 c Heatmap plot reveals the expression of the leading 10 function genes determined in each cluster. d Dot plot reveals the expression of astrocyte gene modules in each significant cell type. 19459027 e 19459028 Dot plot of the enrichR combined rating for the leading enriched GO terms for astrocyte gene modules M12 and M14. 19459027 f WGCNA dendrogram of Cortical Ex gene modules. g 19459028 Dot plot reveals the expression of Cortical Ex gene modules in each significant cell type. 19459027 h 19459028 UMAP plots of module center gene expression rating for Cortical Ex gene modules M1-- 4. i Co-expression plots of the leading 25 module genes for Cortical Ex gene modules M1-- 4. 19459027 j Dot plot of the enrichR combined rating for the leading enriched GO terms for cortical excitatory nerve cell gene modules M1-- 4. Extended Data Fig. 3 Maturation of astrocytes in glia-enriched cortical organoids. 19659230 a 19459028 Immunostaining of five-month-old glia-enriched cortical organoids: GFAP:: GFP AAV-labeled astrocytes (GFP, green), presynaptic blisters (SV2, gray), postsynaptic density (PSD95, magenta). Inset, a bigger view of astrocyte procedure and synapses. Scale bars, 20 μm and 2 μm (inset). b 19459028 Immunostaining of five-month-old glia-enriched cortical organoids: GFAP:: GFP AAV-labeled astrocytes (GFP, green), glutamate transporter (EAAT2, magenta). Scale bar, 20 μm. 19459027 c 19459028 Immunostaining of five-month-old glia-enriched cortical organoids: GFAP:: GFP AAV-labeled astrocytes (GFP, green) and matricellular protein (HEVIN, magenta). Scale bar, 5 μm. 19459027 d Immunostaining of five-month-old glia-enriched cortical organoids: GFAP:: GFP AAV-labeled astrocytes (GFP, green), inward-rectifier potassium channel (Kir4.1, magenta), postsynaptic density protein (PSD95, cyan). Scale bar, 2 μm. e 19459028 Immunostaining of five-month-old glia-enriched cortical organoids: astrocytes (hGFAP, green), Connexin 43 (CX43, magenta). Inset, a bigger view of astrocyte procedures and expression of Connexin 43. Scale bars, 20 μm and 2 μm (inset). 19459027 f 19459028 Metrology of glutamate uptake of neural progenitor cells (NPCs, n= 8) and astrocytes cleansed from five-month-old glia-enriched cortical organoids stemmed from Hues6 (n= 8), iPSC822 (n= 8) and H9 (n= 7) stem cell lines. Each dot represents one independent experiment. Bars, imply ± s.e.m. 19659231 Source information 19659232 Extended Data Fig. 4 Formation of anatomically specified morphological subclasses of human astrocytes in engrafted glia-enriched cortical organoids. 19659233 a Immunostaining of glia-enriched cortical organoid transplants: human nuclear antigen (HuNu, Cyan), astrocytes (GFAP, green), nerve cells (NeuN, magenta). Scale bar, 20 μm. 19459027 b 19459028 Metrology of the portion of NeuN 19459290 + 19459291 HuNu 19459290 + 19459291 cells amongst NeuN + 19459291 cells (left) or huGFAP + GFAP + cells amongst GFAP 19459290 + 19459291 cells (right) in the transplants. n= 4 transplants. Bars, indicate ± s.e.m. c Immunostaining of the center (left) and border (right) of glia-enriched cortical organoid transplants: human astrocytes (hGFAP, green), astrocytes (GFAP, magenta). Scale bars, 50 μm. d Immunostaining of the center (left) and border (right) of glia-enriched cortical organoid transplants: oligodendrocyte progenitor cells (PDGFRα, green), human nuclear antigen (HuNu, magenta). Inset, bigger views of human oligodendrocyte progenitor cells. Scale bars, 50 μm and 10 μm (inset). 19459027 e 19459028 Immunostaining of human interlaminar, protoplasmic and fibrous astrocytes in glia-enriched cortical organoid transplants: human astrocytes (hGFAP, green), white matter (myelin standard protein or MBP, gray). Scale bars, 20 μm. 19459027 f 19459028 Immunostaining of glia-enriched cortical organoid transplants: human cells (GFP, green), astrocytes (S100B, gray; CD44, magenta). Scale bar, 100 μm. 19459027 g 19459028 Immunostaining of the top, center and bottom areas of the transplants: astrocytes (CD44, magenta), white matter (myelin fundamental protein or MBP, gray). Scale bars, 20 μm. 19659234 Source information 19659235 Extended Data Fig. 5 Spatial transcriptome profiling of layer-specific subclasses of human astrocytes in engrafted glia-enriched cortical organoids. 19659236 a Immunofluorescent staining images on the left revealing hGFAP in yellow, SOX9 in magenta and CYTO-13 in blue, in addition to the cell division of SOX9 19459290 + 19459291/ CYTO-13 19459290 + 19459291 Cells portrayed in magenta within a chosen ROI on the. Scale bars, 100 μm. b Box plots portraying the stabilized expression levels of picked genes in each group (pial= 10 ROIs, cortex= 16 ROIs, WM= 12 ROIs). Centerline, mean; box limitations, upper and lower quartiles; hairs, 1.5 × interquartile variety; points, outliers. 19459027 c Bubble plots portraying leading considerable terms determined from GSEA utilizing a weighted Kolmogorov-- Smirnov test. NES, stabilized enrichment rating. Considerable limits set at adjusted p worth < 0.1. 19659237 Extended Data Fig. 6 Close association of human protoplasmic astrocytes with synapses and host vasculature in engrafted glia-enriched cortical organoids. 19659238 a Immunostaining of transplants: human astrocytes (hGFAP, green), glutamate transporters (EAAT2, magenta). Scale bar, 20 μm. 19459027 b 19459028 Immunostaining of transplants: human astrocytes (hGFAP, green), matricellular protein (HEVIN, gray), postsynaptic density (PSD95, magenta). Scale bar, 20 μm. 19459027 c Electron micrograph of a multi-synaptic bouton in an eight-month-old transplant. Arrowheads: synapses. Scale bar, 1 μm. 19459027 d 3D restoration of serial area electron microscopic lense pictures of synaptic structures in an eight-month-old transplant. Presynaptic bouton (orange), postsynaptic density (magenta), dendritic spinal column (blue). Scale bar, 1 μm. 19459027 e Electron micrograph of a synapse within the transplant. Left, presynaptic bouton (orange), postsynaptic density (PSD, magenta), dendritic spinal column (blue) and astrocytic procedures (AP, green). Initial image. Scale bar, 1 μm. f Immunostaining of transplants at 2 (left panel) or 7 months (ideal panel) post-transplantation: capillary (Ly6C, cyan). Scale bars, 100 μm. 19459027 g 19459028 Ly6C + 19459291 location (left) or vessel size (right) in transplants at 2 months (n= 4 transplants) or 7 months (n= 4 transplants) post-transplantation versus contralateral mouse cortex (Ms brain; n= 8 mice). Bars, indicate ± s.e.m. Two-sided t-test, ** p=0.001, **** p < 0.0001. hImmunostaining of transplants: human nuclear antigen (HuNu, green), pericytes (NG2, magenta, suggested by arrowhead in the left image), monocytes (Ly6C, gray, shown by arrowhead in the center image), endothelial cells (CD31, magenta). Scale bars, 10 μm. iImmunostaining of transplants (left) or host brains (right): human nuclear antigen (HuNu, green), microglia (IBA1, magenta). Inset: microglia. Scale bars, 50 μm and 10 μm (insets). jImmunostaining of transplants: human astrocytes (hGFAP, green), inward-rectifier potassium channel (Kir4.1, magenta), capillary (Ly6C, cyan). Scale bar, 20 μm. kImmunostaining of transplants: human astrocytes (hGFAP, green), glucose transporter (Glut1, magenta) and capillary (Ly6C, cyan). Scale bar, 20 μm. lConfocal pictures of the transplant utilized in EM research studies: capillary (DiI, magenta), transplant (GFP, green). Scale bar, 1 mm. m3D restoration of series SEM images: tight junction (yellow) and basement membrane (blue). Scale bar, 1 μm.
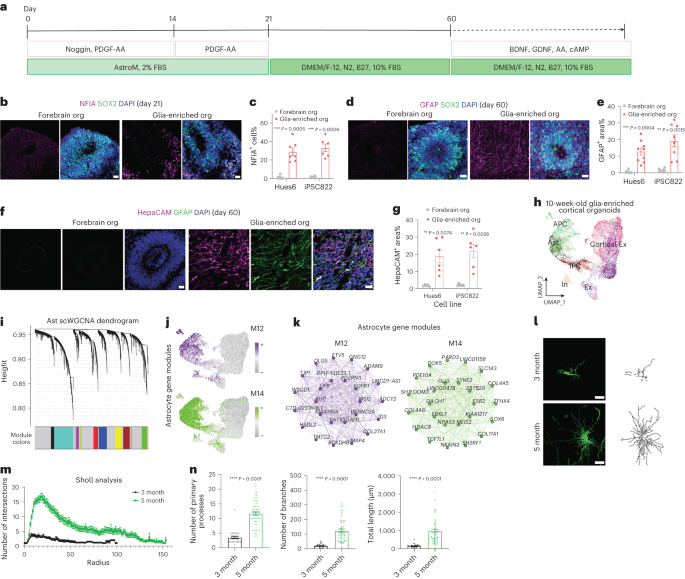
Morphological diversification and functional maturation of human astrocytes in glia-enriched cortical organoid transplanted in mouse brain